Current Research
Microbial host engineering for robust production platforms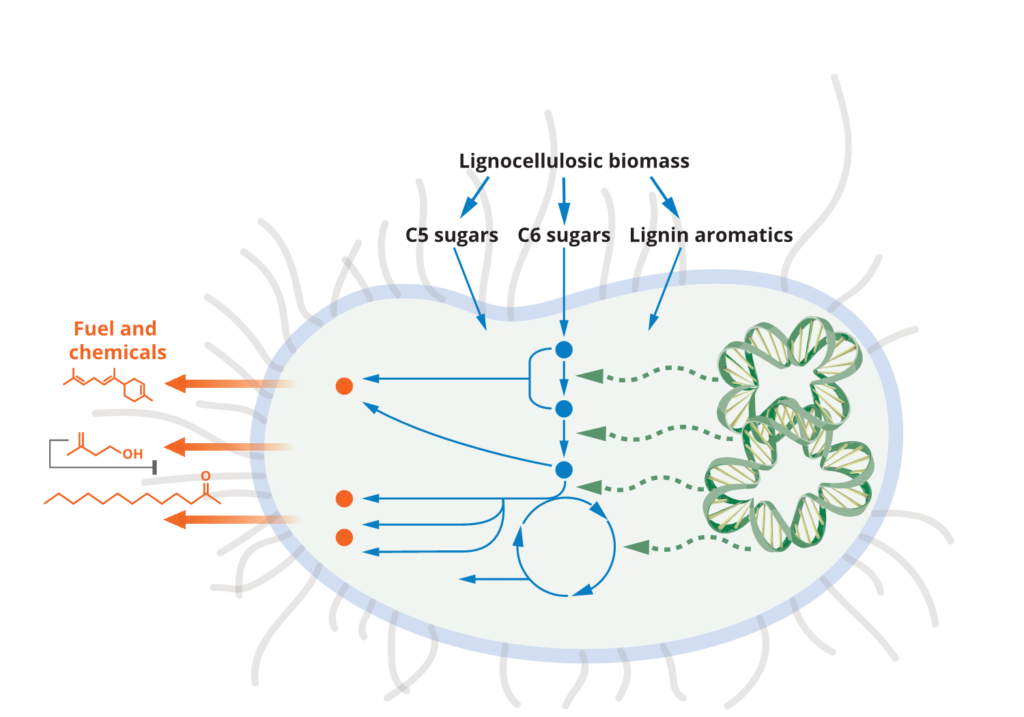
Developing robust microbial systems for bioconversion relies on many aspects beyond the important work of pathway engineering. The toxicity of final products, intermediates and components of a sustainable carbon source is one aspect that requires host engineering. Over the years we have conducted many studies to improve tolerance of the microbial host system to improve and restore production capacities in the presence of such inhibitory compounds. We have also examined routes to improve carbon uptake and utilization in microbial hosts – another aspect of host engineering that is needed for an optimal host. We have approached these challenges from the angle of developing new function in model hosts (e.g. E. coli or S. cerevisiae) or using hosts with native capabilities (E.g P. putida, C. glutamicum, R. toruloides). Most recently, we have initiated studies to better understand strain physiology during scale-up to liter scales in fed-batch modes. We also continue to develop tools for microbial strain engineering that are broadly applicable across multiple bacterial and fungal strains.
Selected Papers:
- Eng T, Banerjee D, Menasalvas J, Chen Y, Gin J, Choudhary H, Baidoo EEK, Chen JH, Ekman A, Kakumanu R, Liu-Diercks Y, Codik A, Larabell C, Gladden J, Simmons BA, Keasling JD, Petzold CJ, Mukhopadhyay A* 2023 Ensemble and Iterative Engineering for Maximized Bioconversion to the Blue Pigment, Indigoidine from Non-Canonical Sustainable Carbon Sources Cell Reports 2023 OA
- Banerjee, D.§, Eng, T.§, Lau, A.K., Sasaki, Y., Wang, B., Chen, Y., Prahl, J.-P., Singan, .R., Herbert, RA., Liu, Y., Tanjore, D., Petzold, CJ., Keasling, JD., Mukhopadhyay, A* 2020. Genome-scale metabolic rewiring improves titers rates and yields of the non-native product indigoidine at scale. Nat. Commun. 11, 5385. 2020 OA
- Wehrs M, Tanjore D, Eng T, Lievense J, Pray TR, Mukhopadhyay A* Engineering robust production microbes for large scale cultivation Trends in Microbiology 2019 OA
- Frederix M, et al Development of an E. coli strain for one-pot biofuel production from ionic liquid pretreated cellulose and switchgrass. Green Chemistry 2016
Signal Transduction and response regulation in bacteria
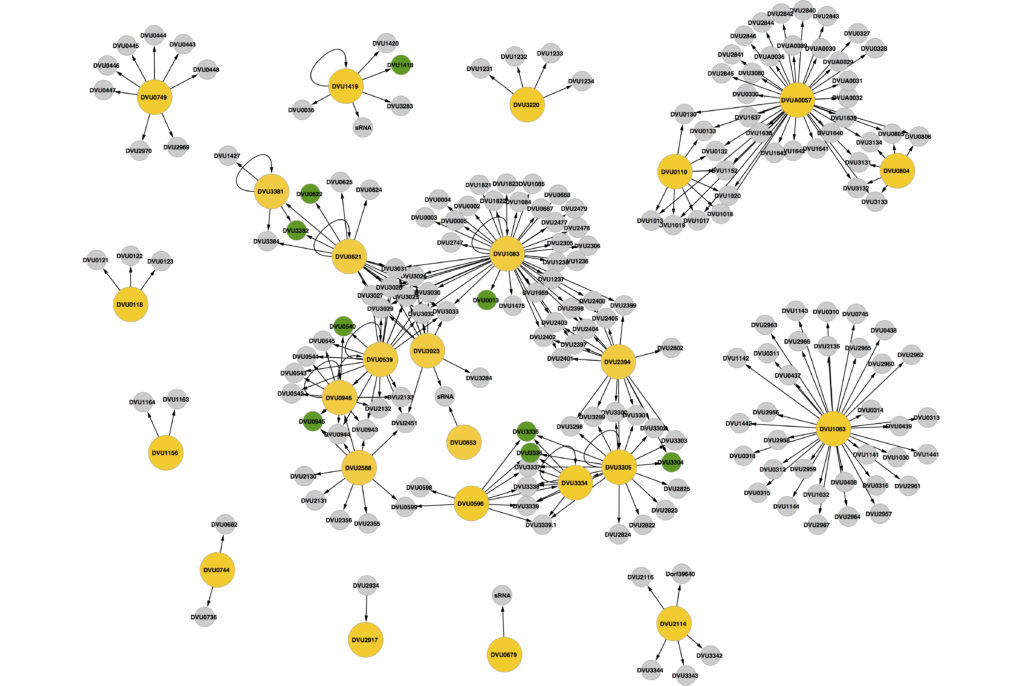 Signaling systems are critical to bacteria in enabling them to continually monitor the environment and respond appropriately to any changes. The numbers and types of signaling systems a microbe possesses is an indication, both the variability of its environment, as well as its ability to perceive and fine-tune its response to diverse signals. We study signaling systems in microbes important for elemental cycling and a variety of environmental impacts, such as nitrate, sulfate and metal homeostasis.
Signaling systems are critical to bacteria in enabling them to continually monitor the environment and respond appropriately to any changes. The numbers and types of signaling systems a microbe possesses is an indication, both the variability of its environment, as well as its ability to perceive and fine-tune its response to diverse signals. We study signaling systems in microbes important for elemental cycling and a variety of environmental impacts, such as nitrate, sulfate and metal homeostasis.
We have examined all two components signaling systems in the model sulfate reducer Desulfovibrio vulgaris Hildenborough. Two component signaling in this organism spans transcriptionally modulated, cyclic-di-GMP modulated as well as chemotaxis modulated via direct protein-protein interactions. To examine transcriptionally acting response regulators in bacteria, we developed the DAP chip method. This enabled the evaluation of all DNA binding RRs in D. vulgaris and revealed unique regulatory clusters and interactions. It also led to the discovery of new motifs and provided the fundamental understanding of all genes that are targeted by two component signaling in this bacterium.
We have now developed a sequencing based DAP-seq to examine signaling in a range of environmentally important organisms. Our recent focus is on denitrifying bacteria such as Pseudomonas stutzeri along with many other isolates from environments such as the Oakridge Field research site.
Selected Papers:
- Garber ME, Frank V, Kazakov AE, Zhang H, Incha MR, Keasling JD, Rajeev L, Mukhopadhyay A* Evolutionarily Driven Domain Swap Alters Sigma Factor Dependence in Bacterial Signaling System mBio 2023
- Rajeev L, et al. Systematic mapping of two component response regulators to gene targets in a model sulfate reducing bacterium. Genome Biology 2011
Discovery of genes on mobile genetic elements from natural isolates
 We have optimized methods to examine native plasmids from natural soil and ground water systems. Examination of genes on these widely present but poorly studied systems reveal striking maintenance of antibiotic and metal resistance genes regardless of similarity of the microbial community. Our samples come from pristine wells that serve as control for the OR FRC S3 waste sites.
We have optimized methods to examine native plasmids from natural soil and ground water systems. Examination of genes on these widely present but poorly studied systems reveal striking maintenance of antibiotic and metal resistance genes regardless of similarity of the microbial community. Our samples come from pristine wells that serve as control for the OR FRC S3 waste sites.
Selected Papers:
Kothari A, Roux S, Zhang H, Prieto A, Soneja D, Chandonia J-M, Spencer S, Wu X, Sara Altenburg, Fields MW, Deutschbauer AM, Arkin AP, Alm EJ, Chakraborty R, Mukhopadhyay A* Ecogenomics of groundwater phages suggests niche differentiation linked to specific environmental tolerance mSystems 2021
Kothari A, Wu Y-W, Chandonia J-M, Charrier M, Rajeev L, Rocha AM, Joyner DC, Hazen TC, Singer SW, Mukhopadhyay A Large circular plasmids from groundwater plasmidomes span multiple incompatibility groups and are enriched in multimetal resistance genes. mBio 2019
Studies of microbial system in the soil in plant microbe systems and for decontamination

We study a range of bacterial systems to examine biological processes in the soil, in plant microbe interfaces and use these to develop solutions to improve soil health and remove toxic contaminations.
Selected Papers:
- Priya et al Assessing horizontal gene transfer in the rhizosphere of Brachypodium distachyon using fabricated ecosystems (EcoFABs) AEM, 2024 OA
Examination of impact of small molecules on microbial, plant microbial and metazoan systems
Selected Papers:
Herbert RA, Eng T, Martinez U, Wang B, Langley S, Wan K, Pidatala V, Hoffman E, Chen JC, Bissell MJ, Brown JB, Mukhopadhyay A* Mortimer JC* Rhizobacteria mediate the phytotoxicity of a range of biorefinery‐relevant compounds Environmental Toxicology and Chemistry 2019 OA
Developing artificial biosynthetic pathways
- Huang J, Quest A, Cruz-Morales P, Deng K, Pereira JH, Van Cura D, Kakumanu R, Baidoo EEK, Dan Q, Chen Y, Petzold CJ, Northen TR, Adams PD, Clark DS*, Balskus EP, Hartwig JF*, Mukhopadhyay A*, Keasling JD* Complete integration of carbene-transfer chemistry into biosynthesis Nature 2023
- Huang J, Liu Z, Bloomer B, Clark DS*, Mukhopadhyay, A*, Keasling JD*, Hartwig JF*. Unnatural Biosynthesis by an Engineered Microorganism with Heterologously Expressed Natural Enzymes and an Artificial Metalloenzyme Nature Chemistry 2021
Completed Research (since 2007)
The microCLeAN G-agent mineralization project
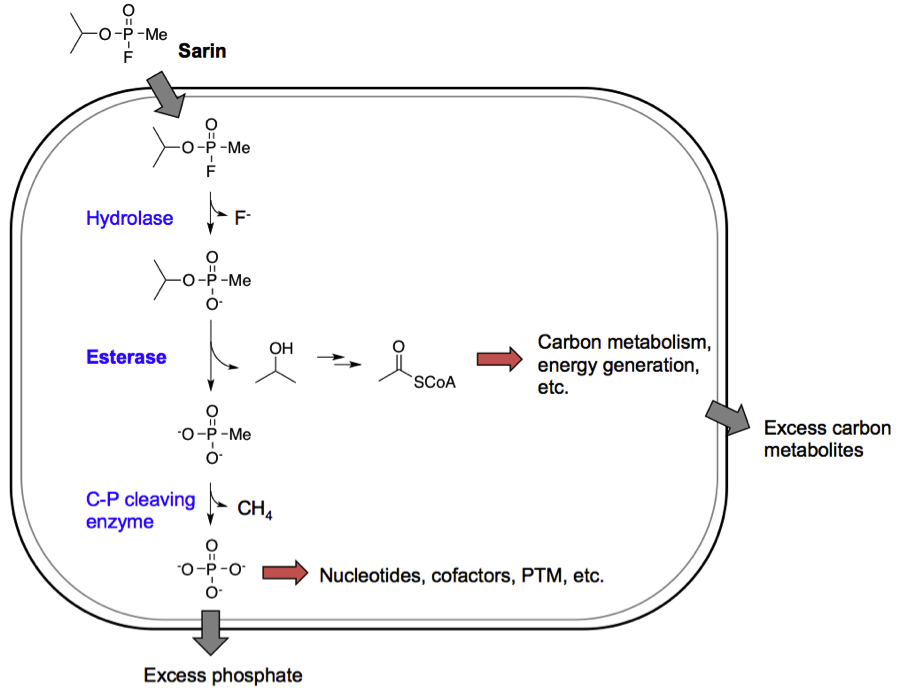 In this DARPA funded project we developed a bacterial platform for the bioremediation of phosphonate ester containing nerve agents. We introduced catabolic enzyme pathways into E. coli to mineralize sarin, a phosphonate ester, into simple phosphorus and carbon units that can be utilized for the organism’s own growth. The mineralization of the carbonaceous part of Sarin is now published. We hope to publish the mineralization of the phosphonate part of soon.
In this DARPA funded project we developed a bacterial platform for the bioremediation of phosphonate ester containing nerve agents. We introduced catabolic enzyme pathways into E. coli to mineralize sarin, a phosphonate ester, into simple phosphorus and carbon units that can be utilized for the organism’s own growth. The mineralization of the carbonaceous part of Sarin is now published. We hope to publish the mineralization of the phosphonate part of soon.
- Brown ME, Mukhopadhyay A, Keasling JD*. Engineering bacteria to catabolize the carbonaceous component of sarin: teaching coli to eat isopropanol. ACS Synthetic Biology 2016
Discovery of Alkane utilization genes
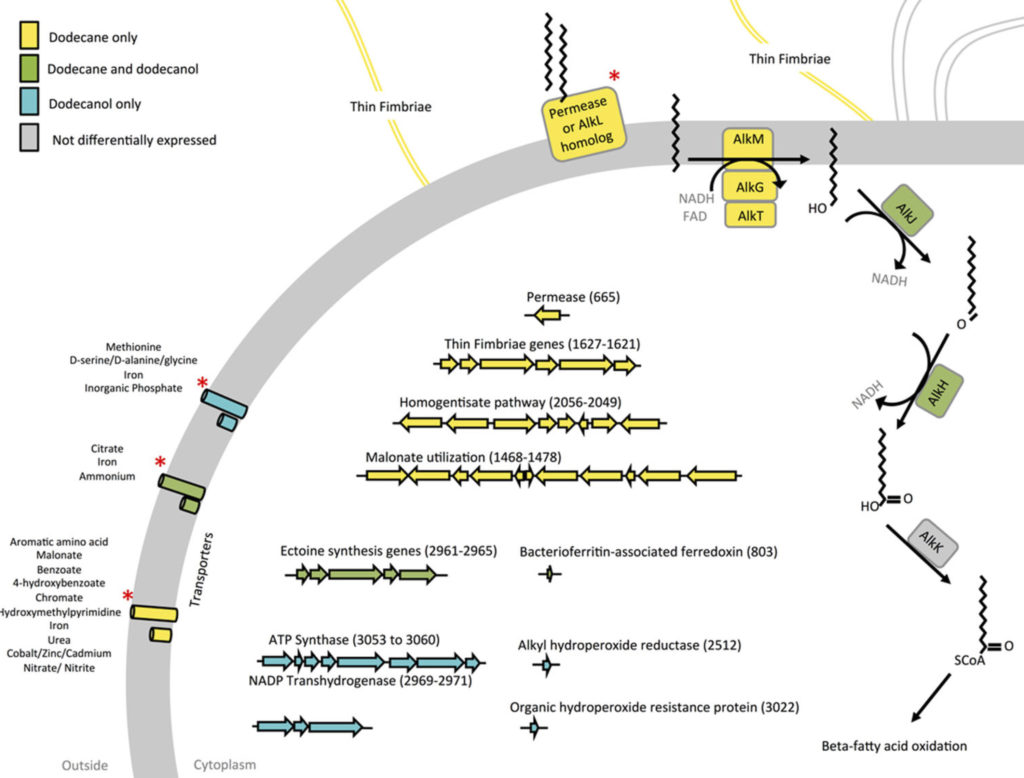 We examined the alkane utilizing ability of a newly sequenced microbe Acinetobacter venetianus RAG1. The RNAseq evaluation of this organism cultivated under various carbon sources relevealed the key genes involved in Alkane import and metabolism
We examined the alkane utilizing ability of a newly sequenced microbe Acinetobacter venetianus RAG1. The RNAseq evaluation of this organism cultivated under various carbon sources relevealed the key genes involved in Alkane import and metabolism
- Kothari A, Charrier M, Wu Y-W, Malfatti S, Zhou CE, Singer SW, Dugan L, Mukhopadhyay A*. Transcriptomic analysis of the highly efficient oil-degrading bacterium Acinetobacter venetianus RAG-1 reveals genes important in dodecane uptake and utilization. FEMS Microbiology Letters 2016
Signaling and gene regulation in dominant cyanobacteria in Desert soil crusts
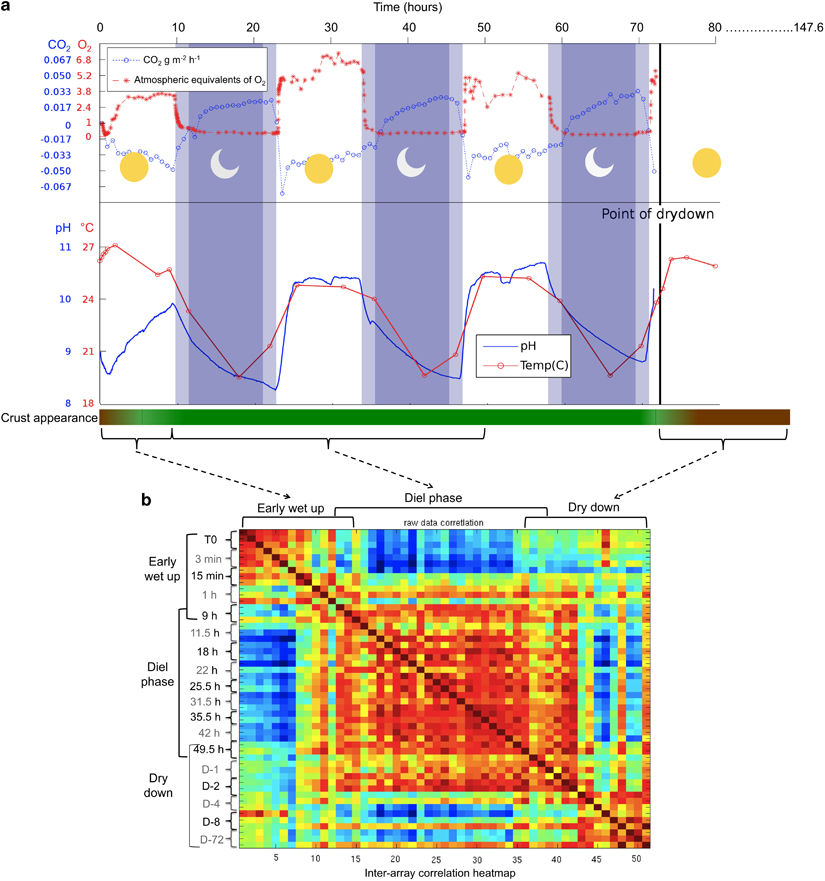 We examined the physiology of Microcoleus vaginatus, the dominant cyanobacerium in desert soil crusts as it emerges from and reenters long term desiccation by simulating rainfall, diurnal cycles and dry-down. Read our paper in the ISME journal.
We examined the physiology of Microcoleus vaginatus, the dominant cyanobacerium in desert soil crusts as it emerges from and reenters long term desiccation by simulating rainfall, diurnal cycles and dry-down. Read our paper in the ISME journal.
Functional Genomics in the model sulfate reducing bacterium Desulfovobrio vulgaris Hildenborough
We conducted a large set of studies for the sulfate reduced D. vulgaris to examine its response to salt, pH, oxygen, nitrate, nitrite as well as its adaptation to several of these stresses, its syntrophic interaction with methanogens and its cellular state in a biofilm.